hotels <- read_csv("data/hotels.csv") |>
mutate(across(where(is.character), as.factor))
count(hotels, children)# A tibble: 2 × 2
children n
<fct> <int>
1 children 4039
2 none 45961Lecture 23
Cornell University
INFO 2951 - Spring 2025
April 17, 2025
ae-21Instructions
ae-21 (repo name will be suffixed with your GitHub name).renv::restore() to install the required packages, open the Quarto document in the repo, and follow along and complete the exercises.recipe()recipe()Creates a recipe for a set of variables
step_*()Adds a single transformation to a recipe. Transformations are replayed in order when the recipe is run on data.
# A tibble: 45,000 × 1
arrival_date
<date>
1 2016-04-28
2 2016-12-29
3 2016-10-17
4 2016-05-22
5 2016-03-02
6 2016-06-16
7 2017-02-13
8 2017-08-20
9 2017-08-22
10 2017-05-18
# ℹ 44,990 more rows# A tibble: 45,000 × 4
arrival_date arrival_date_dow arrival_date_month arrival_date_year
<date> <fct> <fct> <int>
1 2016-04-28 Thu Apr 2016
2 2016-12-29 Thu Dec 2016
3 2016-10-17 Mon Oct 2016
4 2016-05-22 Sun May 2016
5 2016-03-02 Wed Mar 2016
6 2016-06-16 Thu Jun 2016
7 2017-02-13 Mon Feb 2017
8 2017-08-20 Sun Aug 2017
9 2017-08-22 Tue Aug 2017
10 2017-05-18 Thu May 2017
# ℹ 44,990 more rowsstep_holiday() + step_rm()Generate a set of indicator variables for specific holidays.
step_holiday() + step_rm()Rows: 45,000
Columns: 11
$ arrival_date_dow <fct> Thu, Thu, Mon, Sun, Wed, Thu, Mon, Sun, Tue,…
$ arrival_date_month <fct> Apr, Dec, Oct, May, Mar, Jun, Feb, Aug, Aug,…
$ arrival_date_year <int> 2016, 2016, 2016, 2016, 2016, 2016, 2017, 20…
$ arrival_date_AllSouls <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_AshWednesday <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_ChristmasEve <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_Easter <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_ChristmasDay <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_GoodFriday <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_NewYearsDay <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_PalmSunday <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…To predict the outcome of a new data point:
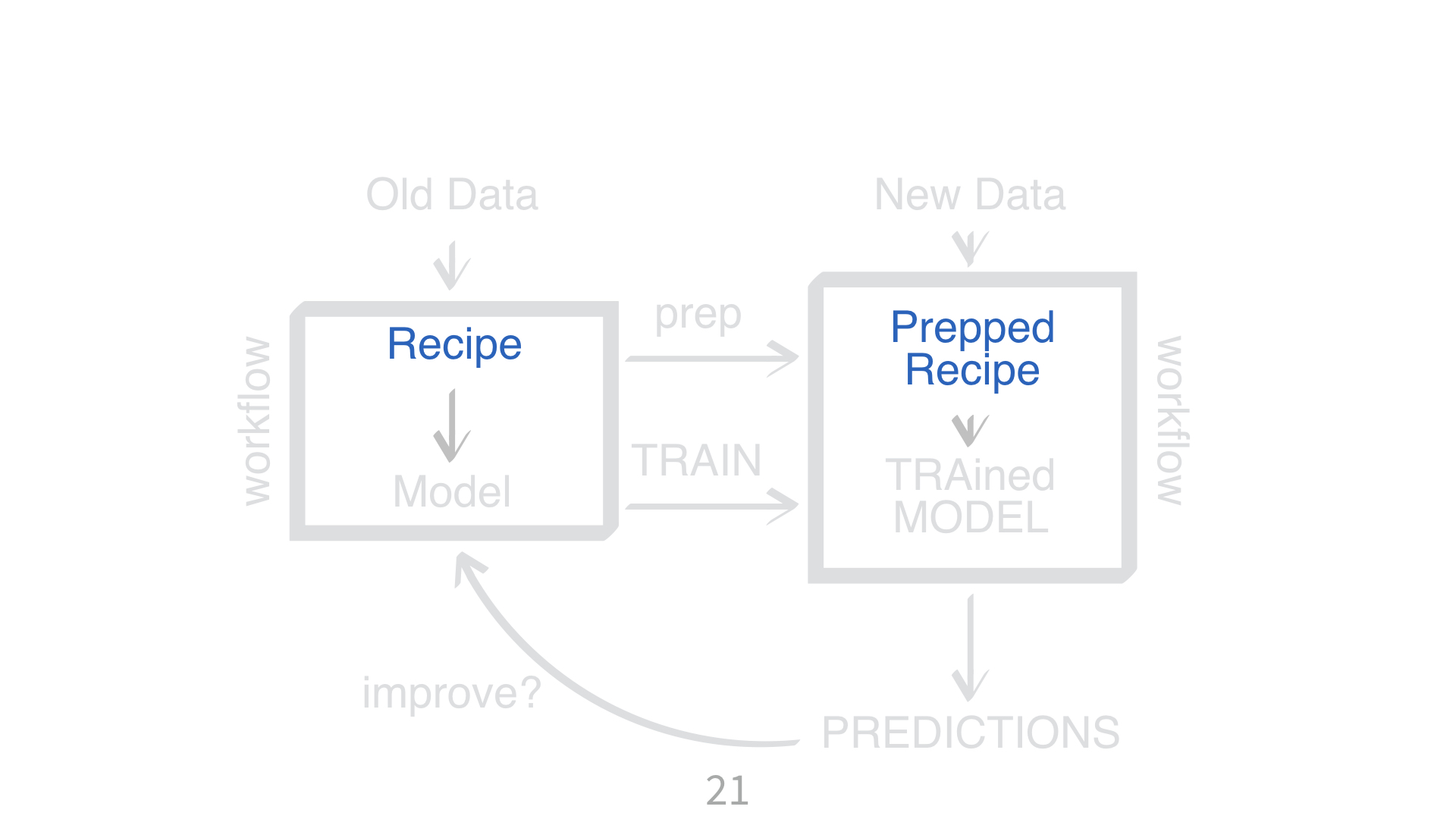
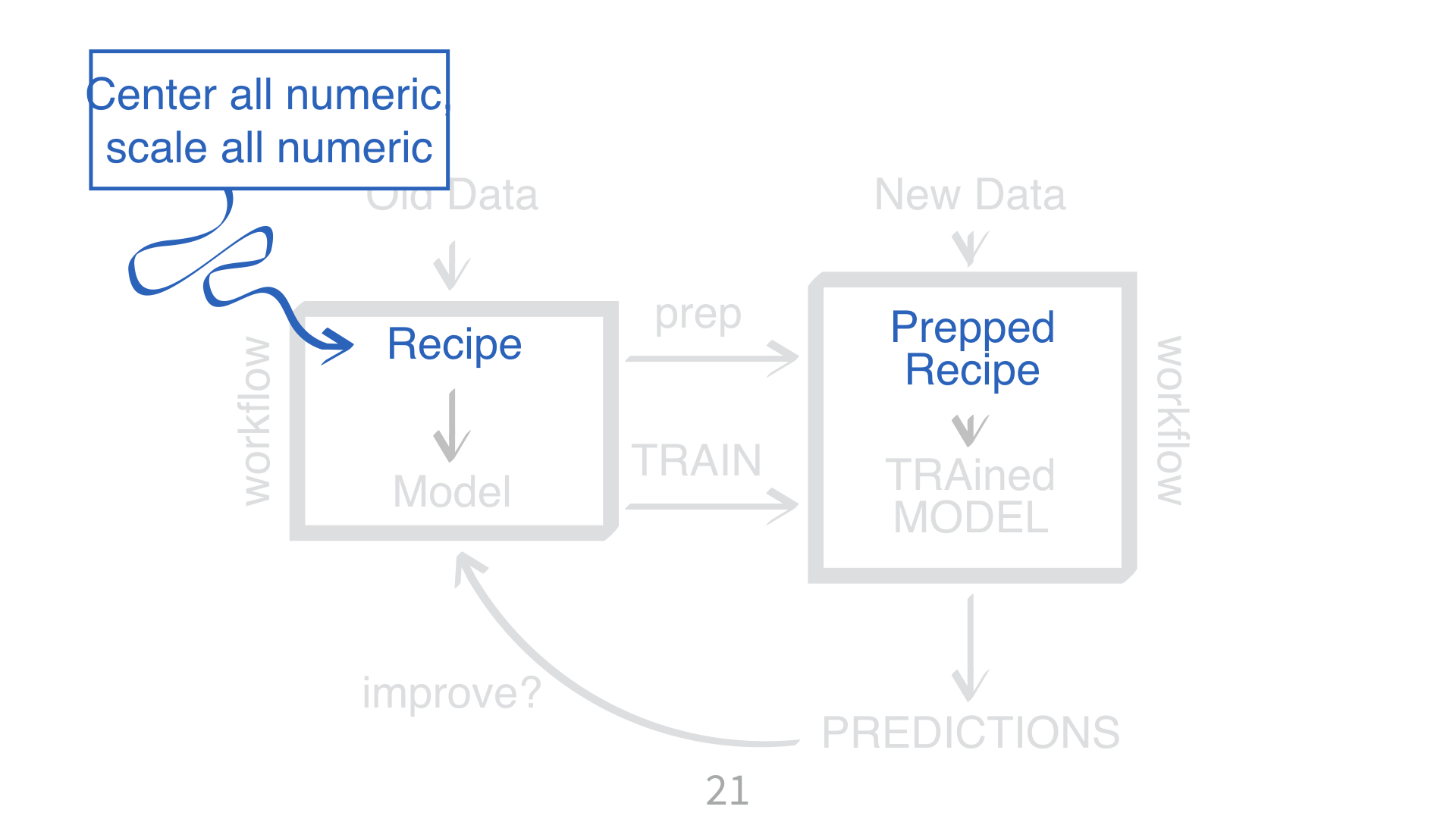
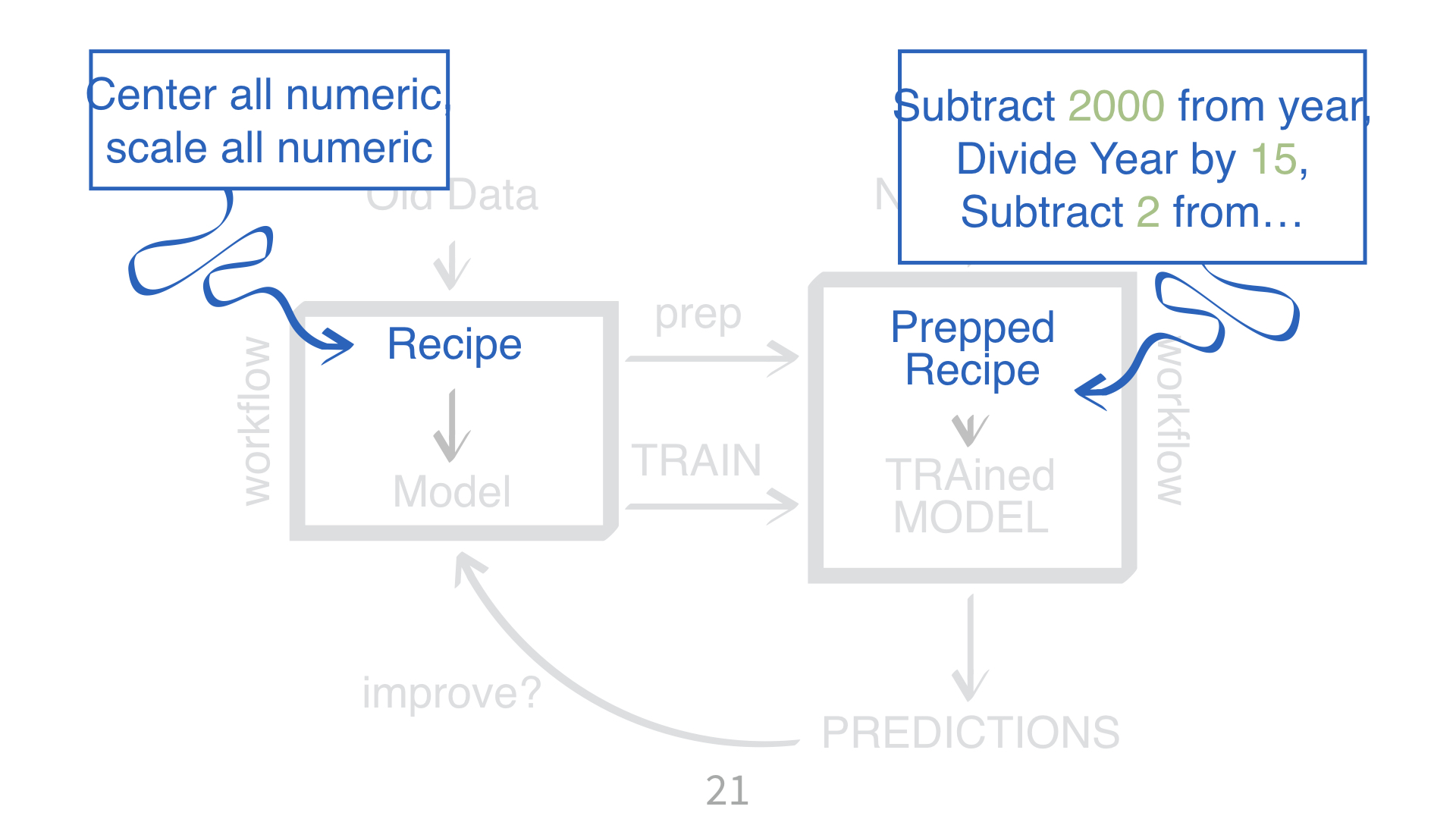
KNN requires all numeric predictors, and all need to be centered and scaled.
What does that mean?
Why do you need to “train” a recipe?
Imagine “scaling” a new data point. What do you subtract from it? What do you divide it by?
# A tibble: 5 × 1
meal
<fct>
1 SC
2 BB
3 HB
4 Undefined
5 FB # A tibble: 50,000 × 5
SC BB HB Undefined FB
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 0 0 0 0
2 0 1 0 0 0
3 0 1 0 0 0
4 0 1 0 0 0
5 0 1 0 0 0
6 0 1 0 0 0
7 0 0 1 0 0
8 0 1 0 0 0
9 0 0 1 0 0
10 1 0 0 0 0
# ℹ 49,990 more rows# A tibble: 5 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 2.38 0.0183 130. 0
2 mealFB -1.15 0.165 -6.98 2.88e-12
3 mealHB -0.118 0.0465 -2.54 1.12e- 2
4 mealSC 1.43 0.104 13.7 1.37e-42
5 mealUndefined 0.570 0.188 3.03 2.47e- 3step_dummy()Converts nominal data into numeric dummy variables, needed as predictors for models like KNN.
How does {recipes} know which variables are numeric and which are nominal?
How does {recipes} know what is a predictor and what is an outcome?
The formula → indicates outcomes vs predictors
The data → is only used to catalog the names and types of each variable
Helper functions for selecting sets of variables
| selector | description |
|---|---|
all_predictors() |
Each x variable (right side of ~) |
all_outcomes() |
Each y variable (left side of ~) |
all_numeric() |
Each numeric variable |
all_nominal() |
Each categorical variable (e.g. factor, string) |
all_nominal_predictors() |
Each categorical variable (e.g. factor, string) that is defined as a predictor |
all_numeric_predictors() |
Each numeric variable that is defined as a predictor |
dplyr::select() helpers |
starts_with(‘NY_’), etc. |
What would happen if you try to normalize a variable that doesn’t vary?
Error! You’d be dividing by zero!
step_zv()Intelligently handles zero variance variables (variables that contain only a single value)
step_normalize()Centers then scales numeric variable (mean = 0, sd = 1)
step_downsample()Instructions
Unscramble! You have all the steps from our knn_rec- your challenge is to unscramble them into the right order!
Save the result as knn_rec
03:00
Source: TMWR
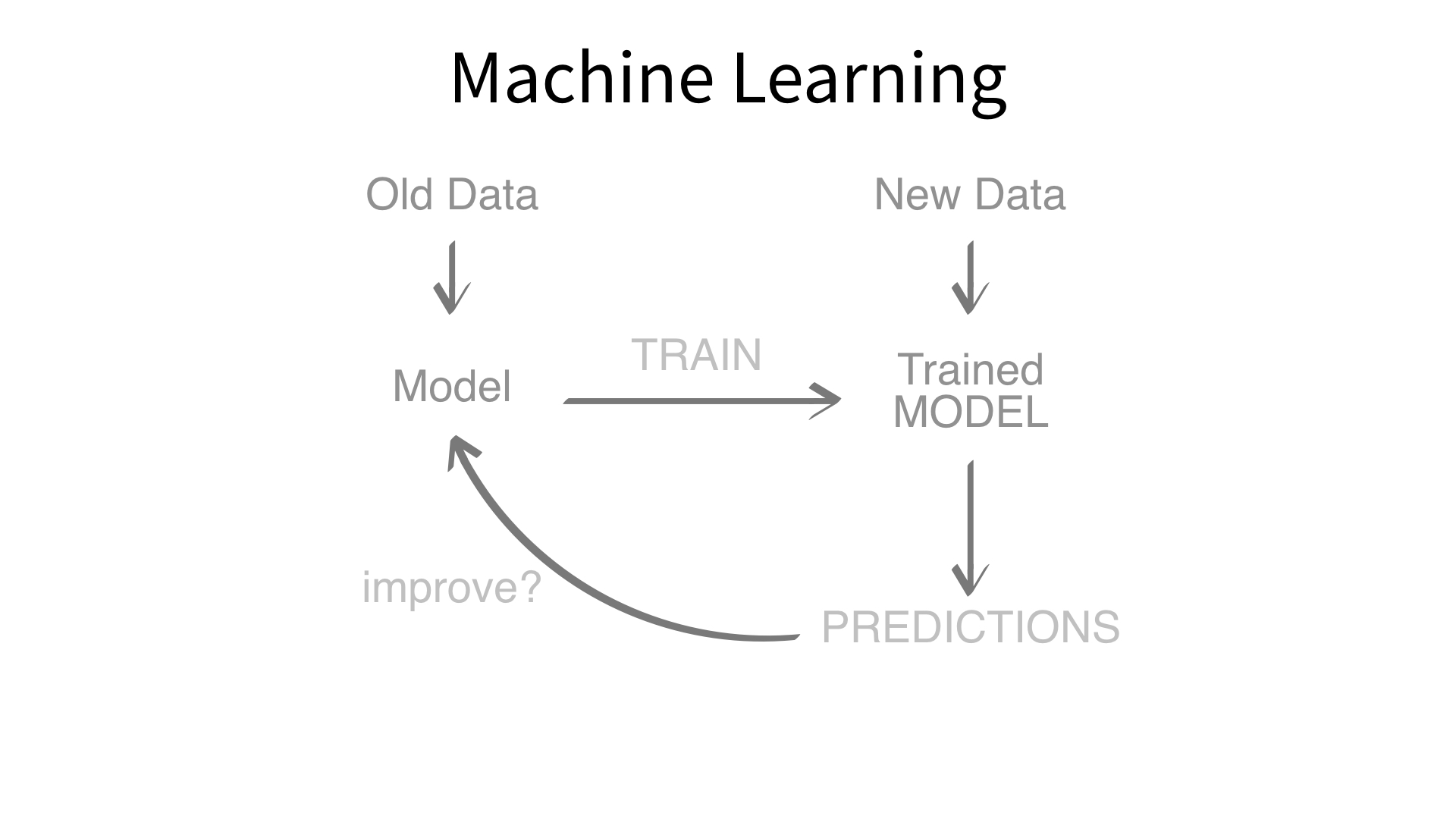
Now we’ve built a recipe.
But, how do we use a recipe?
Feature engineering and modeling are two halves of a single predictive workflow.
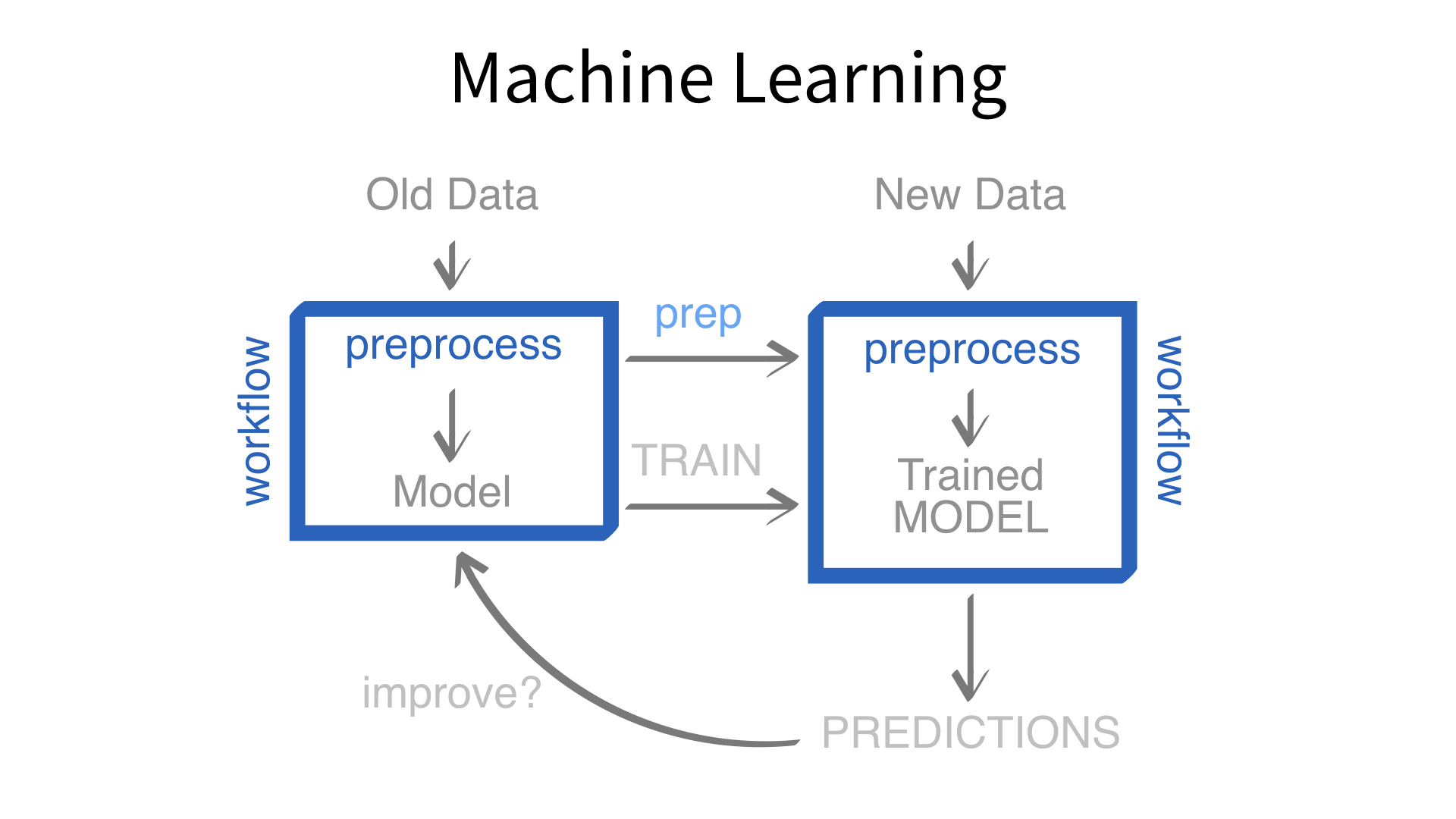
workflow()workflow()Creates a workflow to which you can add a model (and more)
add_formula()Adds a formula to a workflow *
add_model()Adds a parsnip model spec to a workflow
If we use add_model() to add a model to a workflow, what would we use to add a recipe?
Let’s see!
Instructions
Fill in the blanks to make a workflow that combines knn_rec and with knn_mod.
01:00
══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: nearest_neighbor()
── Preprocessor ────────────────────────────────────────────────────────────────
7 Recipe Steps
• step_date()
• step_holiday()
• step_rm()
• step_dummy()
• step_zv()
• step_normalize()
• step_downsample()
── Model ───────────────────────────────────────────────────────────────────────
K-Nearest Neighbor Model Specification (classification)
Computational engine: kknn add_recipe()Adds a recipe to a workflow.
Do you need to add a formula if you have a recipe?
fit()Fit a workflow that bundles a recipe* and a model.
fit_resamples()Fit a workflow that bundles a recipe* and a model with resampling.
Instructions
Run the first chunk. Then try our KNN workflow on hotels_folds. What is the ROC AUC?
03:00
set.seed(100)
hotels_folds <- vfold_cv(hotels_train, v = 10, strata = children)
knn_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.741 10 0.00209 Preprocessor1_Model1
2 brier_class binary 0.173 10 0.00161 Preprocessor1_Model1
3 roc_auc binary 0.830 10 0.00231 Preprocessor1_Model1update_recipe()Replace the recipe in a workflow.
update_model()Replace the model in a workflow.
Instructions
Turns out, the same knn_rec recipe can also be used to fit a penalized logistic regression model. Let’s try it out!
plr_mod <- logistic_reg(penalty = .01, mixture = 1) |>
set_engine("glmnet") |>
set_mode("classification")
plr_mod |>
translate()Logistic Regression Model Specification (classification)
Main Arguments:
penalty = 0.01
mixture = 1
Computational engine: glmnet
Model fit template:
glmnet::glmnet(x = missing_arg(), y = missing_arg(), weights = missing_arg(),
alpha = 1, family = "binomial")03:00
glmnet_wf <- knn_wf |>
update_model(plr_mod)
glmnet_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics() # A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.828 10 0.00218 Preprocessor1_Model1
2 brier_class binary 0.139 10 0.000871 Preprocessor1_Model1
3 roc_auc binary 0.873 10 0.00210 Preprocessor1_Model1workflow() to create explicitly, logical pipelines for training a machine learning model